はじめに
今回は前回の続きです。touch-sp.hatenablog.com
肺の結節影を検出することにチャレンジしました。
前回ダウンロードさせて頂いた154枚の結節影を有する胸部レントゲン写真を使いました。
(学習用:146枚、テスト用:8枚)
非常に少ないですがそれしか手に入らないので仕方がありません。
結果
先に結果を示します。良い結果は得られませんでした。今後の課題です。
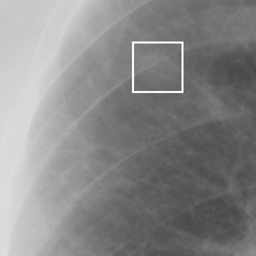
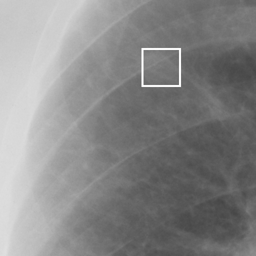
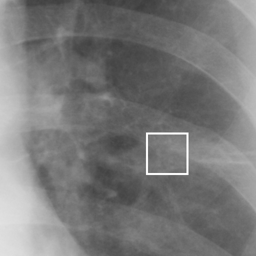
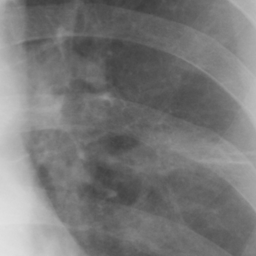
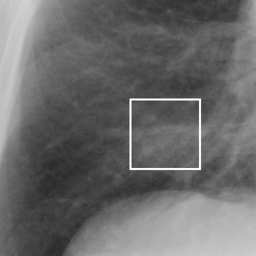
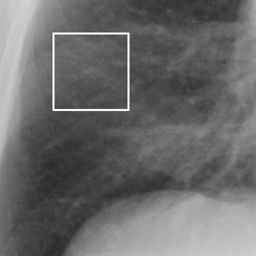
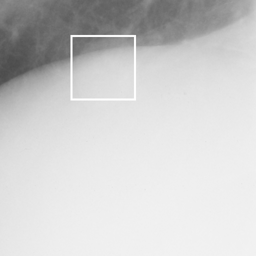
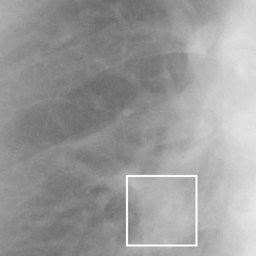
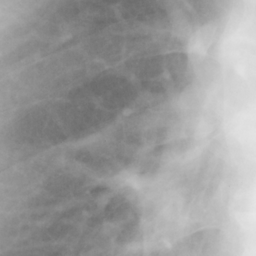
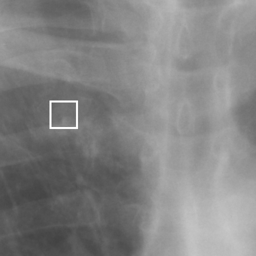
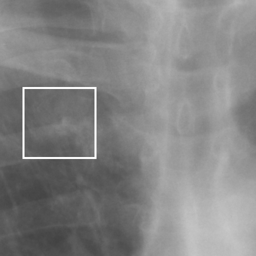
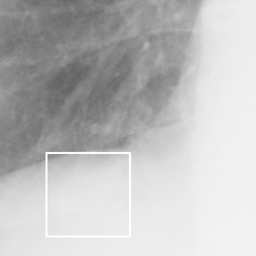
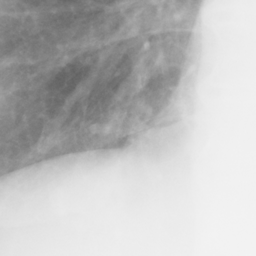
左画像が正解画像、右画像が今回の学習で検出したものです。
8枚のうち正しく検出できたのは2枚だけでした。1枚は違う部位を結節と認識、残りは未検出です。


方法
以下に方法を示します。学習データとテストデータに分ける
from sklearn.model_selection import train_test_split import pandas as pd data = pd.read_csv('CLNDAT_EN.TXT', sep='\t', header=None) train, test = train_test_split(data, train_size=0.95) train.to_csv('train.csv') test.to_csv('test.csv') ''' 読み込む時は train_data = pd.read_csv('train.csv', index_col=0) test_data = pd.read_csv('test.csv', index_col=0) '''
データ標準化のために平均、標準偏差を求める
この部分に限り、今回使用する画像に加えて結節影がうつっていない正常画像93枚も使用しています。import numpy as np import pydicom import glob all_array = [] all_path = glob.glob('./Nodule154images/*.dcm') for path in all_path: a = pydicom.read_file(path) b = a.pixel_array all_array.append(b) all_path = glob.glob('./NonNodule93images/*.dcm') for path in all_path: a = pydicom.read_file(path) b = a.pixel_array all_array.append(b) all_np = np.stack(all_array)/4095 mean = np.mean(all_np) std = np.std(all_np) mean_std = {'mean': mean, 'std': std} import pickle with open('mean_std.pkl', 'wb') as f: pickle.dump(mean_std, f)
Data Augmentation
以下のスクリプトを「grey_augmentation.py」という名前で保存しました。import numpy as np import mxnet as mx from gluoncv.data.transforms import image as timage from gluoncv.data.transforms import bbox as tbbox def random_augmentation(src, label, brightness_delta=512, contrast_low=0.5, contrast_high=1.5, img_mean=0.5, img_std=0.25): """ Note that input image should in original range [0, 255]. Parameters ---------- src : mxnet.nd.NDArray Input image as HWC format. label : numpy.ndarray shape N x 6 (N = number of bbox) brightness_delta : int Maximum brightness delta. Defaults to 32. contrast_low : float Lowest contrast. Defaults to 0.5. contrast_high : float Highest contrast. Defaults to 1.5. Returns ------- mxnet.nd.NDArray Distorted image in HWC format. numpy.ndarray new bounding box """ def brightness(src, delta): if np.random.uniform(0, 1) > 0.5: delta = np.random.uniform(-delta, delta) src += delta src.clip(0, 4095) return src def contrast(src, low, high): if np.random.uniform(0, 1) > 0.5: alpha = np.random.uniform(low, high) src *= alpha src.clip(0, 4095) return src def random_flip(src, label): h, w, _ = src.shape src, flips = timage.random_flip(src, px=0.5, py=0.5) new_bbox = tbbox.flip(label, (w, h), flip_x=flips[0], flip_y=flips[1]) return src, new_bbox src = src.astype('float32') src = brightness(src, brightness_delta) src = contrast(src, contrast_low, contrast_high) src, label = random_flip(src, label) src = src/4095 src = (src-img_mean)/img_std src = src.transpose((2,0,1)) src = mx.nd.concat(src, src, src, dim=0) return src, label
学習
import numpy as np import os import pickle import time import pandas as pd import pydicom import mxnet as mx from mxnet import gluon, autograd from gluoncv import model_zoo from gluoncv.loss import SSDMultiBoxLoss from grey_augmentation import random_augmentation from gluoncv.model_zoo.ssd.target import SSDTargetGenerator target_generator = SSDTargetGenerator( iou_thresh=0.5, stds=(0.1, 0.1, 0.2, 0.2), negative_mining_ratio=-1) ctx = [mx.gpu()] classes = ['nodule'] net = model_zoo.get_model('ssd_512_mobilenet1.0_voc', pretrained=True, ctx = ctx[0], root='./models') net.reset_class(classes) net.hybridize() x = mx.nd.zeros(shape=(1, 3, 512, 512),ctx=ctx[0]) with autograd.train_mode(): _, _, anchors = net(x) anchors = anchors.as_in_context(mx.cpu()) with open('mean_std.pkl', 'rb') as f: mean_std = pickle.load(f) img_mean = mean_std['mean'] img_std = mean_std['std'] csv_data = pd.read_csv('train.csv', index_col=0) def make_train_dataset(): img_list = [] cls_list = [] box_list = [] for i in range(len(csv_data)): file_name = os.path.splitext(csv_data.iloc[i,0])[0] + '.dcm' img = pydicom.read_file(os.path.join('Nodule154images', file_name)) img = img.pixel_array position_x = csv_data.iloc[i,5] position_y = csv_data.iloc[i,6] size = csv_data.iloc[i,2] nodule_size = int(size/(0.175*2)) x_min = position_x - nodule_size x_max = position_x + nodule_size y_min = position_y - nodule_size y_max = position_y + nodule_size if position_x < 1024: crop_xmin = np.random.randint(max(3, x_max-450), x_min-3) else: crop_xmin = np.random.randint(x_max+3, min(2044, x_min+450)) - 512 if position_y < 1024: crop_ymin = np.random.randint(max(3, y_max-450), y_min-3) else: crop_ymin = np.random.randint(y_max+3, min(2044, y_min+450)) - 512 x_min -= crop_xmin x_max -= crop_xmin y_min -= crop_ymin y_max -= crop_ymin img = img[crop_ymin:crop_ymin+512, crop_xmin:crop_xmin+512] img = mx.nd.array(img).expand_dims(2) label = np.array([x_min, y_min, x_max, y_max, 0, 0]).reshape((1, -1)).astype('float32') img, label = random_augmentation(img, label, img_mean=img_mean, img_std=img_std) gt_bboxes = mx.nd.array(np.expand_dims(label[:,:4], 0)) gt_ids = mx.nd.array(np.expand_dims(label[:,4:5], 0)) cls_targets, box_targets, _ = target_generator(anchors, None, gt_bboxes, gt_ids) img_list.append(img) cls_list.append(cls_targets[0]) box_list.append(box_targets[0]) return img_list, cls_list, box_list #hyperparameters train_epochs = 100 train_batch = 4 num_workers = 0 mbox_loss = SSDMultiBoxLoss() ce_metric = mx.metric.Loss('CrossEntropy') smoothl1_metric = mx.metric.Loss('SmoothL1') trainer = gluon.Trainer( net.collect_params(), 'sgd', {'learning_rate': 0.001, 'wd': 0.0005, 'momentum': 0.9}) for epoch in range(train_epochs): img_list, cls_list, box_list = make_train_dataset() epoch_dataset = gluon.data.dataset.ArrayDataset(img_list, cls_list, box_list) train_loader = gluon.data.DataLoader(epoch_dataset, batch_size=train_batch, shuffle=True, last_batch='rollover', num_workers=num_workers) ce_metric.reset() smoothl1_metric.reset() tic = time.time() btic = time.time() for i, batch in enumerate(train_loader): batch_size = batch[0].shape[0] data = gluon.utils.split_and_load(batch[0], ctx_list=ctx, batch_axis=0) cls_targets = gluon.utils.split_and_load(batch[1], ctx_list=ctx, batch_axis=0) box_targets = gluon.utils.split_and_load(batch[2], ctx_list=ctx, batch_axis=0) with autograd.record(): cls_preds = [] box_preds = [] for x in data: cls_pred, box_pred, _ = net(x) cls_preds.append(cls_pred) box_preds.append(box_pred) sum_loss, cls_loss, box_loss = mbox_loss( cls_preds, box_preds, cls_targets, box_targets) autograd.backward(sum_loss) trainer.step(1) ce_metric.update(0, [l * batch_size for l in cls_loss]) smoothl1_metric.update(0, [l * batch_size for l in box_loss]) name1, loss1 = ce_metric.get() name2, loss2 = smoothl1_metric.get() if i % 20 == 0: print('[Epoch {}][Batch {}], Speed: {:.3f} samples/sec, {}={:.3f}, {}={:.3f}'.format( epoch, i, batch_size/(time.time()-btic), name1, loss1, name2, loss2)) net.save_parameters('ssd_512_mobilenet1.0_nodule_%d.params'%train_epochs)
テストデータを変形
テストデータはモデルに代入するために変形が必要です。縮小ではなく切り取りを行いました。切り取りの際には結節影が必ず含まれるように工夫しています。何度も使えるようにpickleで保存しています。import numpy as np import os import pickle import pandas as pd import pydicom csv_data = pd.read_csv('test.csv', index_col=0) def make_test_dataset(): original_img_list = [] test_label_list = [] for i in range(len(csv_data)): file_name = os.path.splitext(csv_data.iloc[i,0])[0] + '.dcm' img = pydicom.read_file(os.path.join('Nodule154images', file_name)) img = img.pixel_array position_x = csv_data.iloc[i,5] position_y = csv_data.iloc[i,6] size = csv_data.iloc[i,2] nodule_size = int(size/(0.175*2)) x_min = position_x - nodule_size x_max = position_x + nodule_size y_min = position_y - nodule_size y_max = position_y + nodule_size if position_x < 1024: crop_xmin = np.random.randint(max(3, x_max-450), x_min-3) else: crop_xmin = np.random.randint(x_max+3, min(2044, x_min+450)) - 512 if position_y < 1024: crop_ymin = np.random.randint(max(3, y_max-450), y_min-3) else: crop_ymin = np.random.randint(y_max+3, min(2044, y_min+450)) - 512 x_min -= crop_xmin x_max -= crop_xmin y_min -= crop_ymin y_max -= crop_ymin img = img[crop_ymin:crop_ymin+512, crop_xmin:crop_xmin+512] label = np.array([x_min, y_min, x_max, y_max]) original_img_list.append(img) test_label_list.append(label) return original_img_list, test_label_list original_img_list, label_list = make_test_dataset() with open('test_data.pkl', 'wb') as f: pickle.dump([original_img_list, label_list], f)
テストデータに対してモデルを適用
import os import pickle import mxnet as mx from PIL import Image, ImageDraw from gluoncv import model_zoo os.mkdir('result') ctx = mx.gpu() classes = ['nodule'] net = model_zoo.get_model('ssd_512_mobilenet1.0_voc', pretrained=True, ctx = ctx, root='./models') net.reset_class(classes) net.load_parameters('ssd_512_mobilenet1.0_nodule.params') net.hybridize() with open('mean_std.pkl', 'rb') as f: mean_std = pickle.load(f) with open('test_data.pkl', 'rb') as f: test_dataset = pickle.load(f) img_mean = mean_std['mean'] img_std = mean_std['std'] for i in range(len(test_dataset[0])): original_img = test_dataset[0][i] label = test_dataset[1][i] src = mx.nd.array(original_img).expand_dims(2) src = src/4095 src = (src-img_mean)/img_std src = src.transpose((2,0,1)) src = mx.nd.concat(src, src, src, dim=0) #正解表示 img = Image.fromarray((original_img/16).astype('uint8'), mode='L') draw = ImageDraw.Draw(img) draw.rectangle(list(label), outline=255, width=4) img.save(os.path.join('result', 'correct_position_%d.png'%i)) #検出結果表示 normalize_img = src.expand_dims(0) class_IDs, scores, bounding_boxs = net(normalize_img.as_in_context(ctx)) result_num = int(mx.nd.sum(scores>0.7).asscalar()) img = Image.fromarray((original_img/16).astype('uint8'), mode='L') draw = ImageDraw.Draw(img) for x in range(result_num): draw.rectangle([int(x.asscalar()) for x in bounding_boxs[0][x]], outline=255, width=4) img.save(os.path.join('result', 'predict_reslt_%d.png'%i))
考察
今回良い結果は得られませんでした。もう少しどうにかなるのではないかと考えています。
この課題にもう少し取り組んでいこうと思います。いつか良い結果がでればまた記事にします。